Abstract
Sideroblastic anemias consist of a heterogeneous group of inherited and acquired disorders. The most common hereditary type is X-linked sideroblastic anemia (XLSA), which is associated with mutations in the erythroid-specific δ-aminolevulinic acid synthase (ALAS2) gene. Heme synthesis starts with the polymerization of glycine and succinyl CoA polymerization and synthesis of δ-aminolevulinic acid (ALA) in the mitochondria. ALAS2 encodes the enzyme that catalyzes these first steps in the heme synthetic pathway in erythroid cells, steps that require pyridoxal 5'-phosphate (PLP) as a cofactor. It has been found that treatment with PLP is effective for a small fraction of XLSA patients, but there are no effective treatments for the other fraction. The aim of this study is to explore the molecular mechanisms of XLSA and to develop new effective therapies.
We used episomal methods to generate induced pluripotent stem cells (iPSCs) from peripheral blood mononuclear cells (PBMCs) of three late-onset XLSA female patients in one family. The cells harbored the heterozygous mutation (R227C) in the ALAS2 gene. Because ALAS2 is located in the X-chromosome, either wild-type or mutant ALAS2 gene is inactivated in the erythroid cells of female heterozygotes. All three patients showed severe anemia and their PBMCs showed skewed X-chromosome inactivation with preferential inactivation of the X chromosome carrying wild-type ALAS2, indicating a condition associated with unbalanced lyonization. From each patient, we successfully established iPSC lines with the active mutant ALAS2 allele and with the active wild-type ALAS2 allele.
We assessed the hematopoietic differentiation potential of these two types of iPSC lines derived from the same patient. Differentiation into hematopoietic progenitor cells (HPCs) using embryoid body formation was comparable in the two groups. However, further differentiation in erythroid culture was significantly impaired in iPSC lines harboring the active mutant ALAS2 allele compared with those harboring the active wild-type ALAS2 allele (CD235a+ cells: 59.20±12.16% with the active wild-type ALAS2 allele vs. 3.95±4.71% with the active mutant ALAS2 allele, p<0.01). Only mutant ALAS2 expression was observed in erythroid cells differentiated from iPS cells harboring the active mutant ALAS2 allele, and only wild-type ALAS2 expression was observed in erythroid cells differentiated from iPS cells harboring the active wild-type ALAS2 allele. Hematopoietic maturation capacity was assessed by performing colony-forming unit (CFU) assays of HPCs (CD34+CD38-CD43+lineage marker-) from iPSC lines derived from the same XLSA patient. Erythroid colony count was significantly less in HPCs from iPSC lines with the active mutant ALAS2 allele, but there was no difference in total colony count between the two types of iPSC lines (erythroid colony numbers: 9.66±10.69 vs. 0±0 per 7,500 HPCs, p<0.01; mixed erythroid colony numbers: 15.00±11.26 vs. 0.66±0.57 per 7,500 HPCs, p<0.01; HPCs with the active wild-type ALAS2 allele vs. HPCs with the active mutant ALAS2 allele).
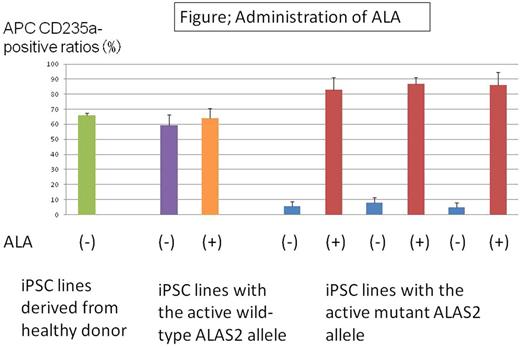
We examined the effect of ALA on the erythroid differentiation of the HPCs. The CD235a-positive erythroid cell ratio of HPCs with the active wild-type ALAS2 allele did not increase following administration of ALA. By contrast, the ratio reached normal levels for HPCs with the active mutant ALAS2 allele (CD235a+ cells: 6.10± 5.61% vs. 85.34± 11.05%, p<0.01; without vs. with administration of ALA).
Our data suggest that our iPSC-based system could be useful for studying the precise molecular mechanisms of XLSA and drug testing.
Morimoto: Grant-in-Aid for JSPS Research Fellow: Research Funding. Takaori-Kondo: celgene: Honoraria, Research Funding; Bristol-Myers Squibb, Novartis, Janssen pharma, Pfizer: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal